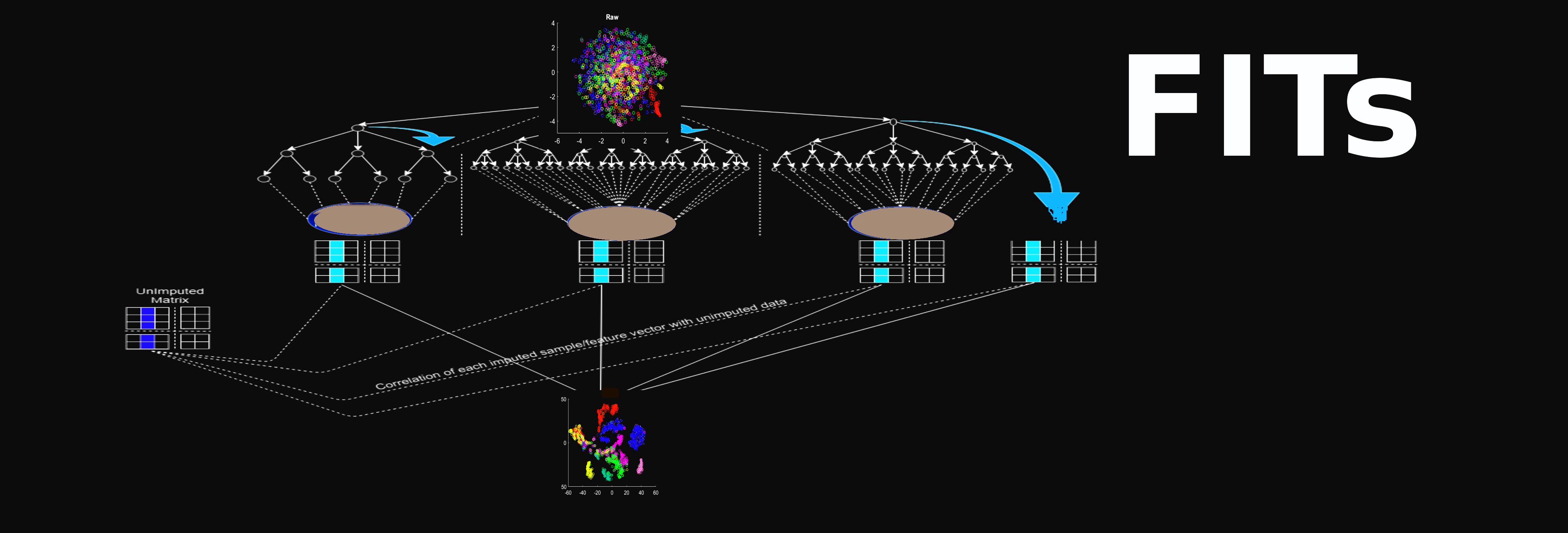
Jump to Matlab_code_help
Other codes : using UniPath and chromatin interaction prediction after application of FITs
Using standalone executable for Linux
After downloading the file FITS.tar.gz gunzip and untar the file as such
$ gunzip FITS.tar.gz
$ tar -xvf FITS.tar
Now include the FITS folder in your PATH
$ export PATH=path_of_FITS_folder:$PATH
Now you can access commands of FITS from any folder.
For execution you have to pass filename of read-counts csv as a input file. Read-count csv file does not contain header and genomic location i.e. it consist of only data on which imputation is going to perform. Row represents sites and column represent samples/cells in csv file.
To get final imputed matrix, you run two phases1 of FITs using command
$ run_FITSPhase1.sh input=unimputed.csv output=imputed.csv
It is not necessary to use any option, however for faster executation you could use option
maxLevel= 2
maxLevel can be between 2 to 6 but not less than 2, such as
$ run_FITSPhase1.sh input=unimputed.csv maxLevel=2 output=imputed.csv
run phase1 of FITS many times, as such
$ run_FITSPhase1.sh input=unimputed.csv output=imputed.csv
$run_FITSPhase1.sh input=unimputed.csv output=imputed.csv
$run_FITSPhase1.sh input=unimputed.csv output=imputed.csv
....
$run_FITSPhase1.sh input=unimputed.csv output=imputed.csv
after executing Phase-1 many times, run the phase-2 of FITs
$run_FITSPhase2.sh input=unimputed.csv output=imputed.csv
For large
read-count matrices
For imputing large read-count matrices without comsuming too much memory on computer use FITSPhase1L and FITSPhase2L
Such that you run FITSPhase1L many times ( more than 5 times) before you finally call FITSPhase2L
$ run_FITSPhase1L.sh input=unimputed.csv output=imputed.csv
$run_FITSPhase1L.sh input=unimputed.csv output=imputed.csv
$run_FITSPhase1L.sh input=unimputed.csv output=imputed.csv
……………………..
run_FITSPhase1L.sh input=unimputed.csv output=imputed.csv
Then run FITSPhase2L in same folder
$ run_FITSPhase1L.sh input =unimputed.csv output=imputed.csv
Using Matlab source code
"# FITS Matlab version"
You have to download matlab code in your local machine/server. For execution you have to pass filename of read-counts csv as a input file. Read-count csv file does not contain header and genomic location i.e. it consist of only data on which imputation is going to perform. Row represents sites and column represent samples/cells in csv file.
Start matlab and
Addpath of FITs using the command
>addpath(path_of_FITs_folder) ;
Now run phase1 of FITs on matlab console
> FITSPhase1 input='<csv file name>'
e.g.
> FITSPhase1 input='sce5_raw.csv'
'sce5_raw.csv' consist of epignome data corresponding to five cell type.
Other optional input parameter you can pass in phase1
>FITSPhase1 input='<csv file name>' output='<name to save imputed file>' maxLevel =<Depth upto which tree will grow>
By default maxLevel set to 4 and output set to 'FITSOutput'.
You can run FITSPhase1 parallely in background using
nohup matlab -nodisplay -nosplash -r "try FITSPhase1 input='<csv file name>'; catch; end; quit" > <name>.txt &
```
You can create n number of imputed matrix generated through phase1. Each run will generate imputed matrix.
Once Phase1 is over then run Phase2 to generate final imputed matrix based on matrix received as output from Phase1. You can run Phase2 on matlab console
> FITSPhase2 input='<csv file name>'
e.g.
> FITSPhase2 input='sce5_raw.csv'
You have to pass same input file as you passed in Phase1. Don't worry Phase2 takes only one minute to generate final output :)
Other optional input parameter you can pass in phase2
Ø FITSPhase2 input='<csv file name>' output='<name to save imputed file same as Phase1>' k =<topk correlated matrix feature/sample value use for final imputation> feature=<1/0 takes values either 1 or 0>
```
Default value for feature is zero. At value 0 phase2 will compute correlation among samples/cell (preffered) while value 1 will compute correlation features/sites wise.
For large
read-count matrices
For imputing large
read-count matrices without comsuming too much memory
on computer use FITSPhase1L and FITSPhase2L
Such that you run
FITSPhase1L many times ( atleast 5 times) before you finally call FITSPhase2L
Run FITSPhase1L many times before calling FITSPhase2L
FITSPhase1L input=unimputed.csv output=imputed.csv
Then run
FITSPhase2L in same folder
$FITSPhase2L input=unimputed.csv output=imputed.csv
As FITSPhase1L occupies less RAM memory you could run multiple processes parralely using nohup
nohup matlab -nodisplay -nosplash FITSPhase1L input='<csv
file name>’ output=’<imputed.csv>'
; > <name>.txt &
nohup matlab -nodisplay -nosplash -r FITSPhase1L input='<csv
file name> ’ output=’<imputed.csv>'
; > <name>.txt &
.....
nohup matlab -nodisplay
-nosplash -r "try FITSPhase1L input='<csv
file name>’ output=’<imputed.csv>'
; catch; end; quit" > <name>.txt &
Then run FITSPhase2L
nohup matlab -nodisplay
-nosplash -r "try FITSPhase2L input='<csv
file name>’ output=’<imputed.csv>'
; catch; end; quit" > <name>.txt &
Using Python code
One needs to have python 3.0+ installed in their machine. You have to download python code in your local machine/server. For execution you have to pass filename of read-counts csv as a input file. Read-count csv file does not contain header and genomic location i.e. it consist of only data on which imputation is going to perform. Row represents sites and column reprent samples/cells in csv file.
$ python3 FITSPhase1.py -i <csv file name>
e.g.
$python3 FITSPhase1.py -i sce5_raw.csv
'sce5_raw.csv' consist of epignome data corresponding to five cell type.
Other optional input parameter you can pass in phase1
$ python3 FITSPhase1.py -i <csv file name> -o <name to save imputed file> -l <Depth upto which tree will grow>
Usage help can be availed by following command
$ python3 FITSPhase1.py -h
By default -l (maxLevel) set to 4 and -o (output) set to 'FITS_OUTPUT'.
You can run FITSPhase1 parallely in background using :
$ nohup python3 FITSPhase1.py -i <csv file name> > <name>.txt &
You can create n number of imputed matrix generated through phase1. Each run will generate imputed matrix. Once Phase1 is over then run Phase2 to generate final imputed matrix based on matrix received as output from Phase1.
$python3 FITSPhase2.py -i <csv file name>
e.g.
$python3 FITSPhase2.py -i sce5_raw.csv
You have to pass same input file as you passed in Phase1. Don't worry Phase2 takes only one minute to generate final output :)
Other optional input parameter you can pass in phase2
$python3 FITSPhase2.py -i <csv file name> -o <output> -t <topk > -c <1/0 takes values either 1 or 0>
Here topk represents number of top correlated vectors to us to build final matrix.
Default value for feature is zero. At value 0 phase2 will compute correlation among samples/cell (preffered) while value 1 will compute correlation features/sites wise.
Usage help can be availed by following command
$ python3 FITSPhase2.py -h
For large
read-count matrices
For imputing large
read-count matrices without consuming too much memory on computer use
FITSPhase1L and FITSPhase2L
Such that you run
FITSPhase1L many times (5 times) before you finally call FITSPhase2L
Run FITSPhase1L
many times before calling FITSPhase2L
nohup python3 FITSPhase1.py -i <csv file name> > <name>.txt &
Then run FITSPhase2L in same folder
$python3 FITSPhase2L.py input -i unimputed.csv -o imputed.csv